# PartSeg

[](https://badge.fury.io/py/PartSeg)
[](https://anaconda.org/conda-forge/partseg)
[](https://pypi.org/project/partseg)
[](https://partseg.readthedocs.io/en/latest/?badge=latest)
[](https://dev.azure.com/PartSeg/PartSeg/_build/latest?definitionId=1&branchName=develop)
[](https://zenodo.org/badge/latestdoi/166421141)
[](https://doi.org/10.1186/s12859-021-03984-1)
[](https://github.com/4DNucleome/PartSeg/blob/master/License.txt)
[](https://github.com/pre-commit/pre-commit)
[](https://github.com/psf/black)

[](https://lgtm.com/projects/g/4DNucleome/PartSeg/alerts/)
[](https://lgtm.com/projects/g/4DNucleome/PartSeg/context:python)
[](https://www.codacy.com/gh/4DNucleome/PartSeg/dashboard?utm_source=github.com&utm_medium=referral&utm_content=4DNucleome/PartSeg&utm_campaign=Badge_Grade)
[](https://codecov.io/gh/4DNucleome/PartSeg)
[](https://deepsource.io/gh/4DNucleome/PartSeg/?ref=repository-badge)
PartSeg is a GUI and a library for segmentation algorithms. PartSeg also provide napari plugins for IO and labels measurement.
This application is designed to help biologist with segmentation based on threshold and connected components.
## Tutorials
- Tutorial: **Chromosome 1 (as gui)** [link](https://github.com/4DNucleome/PartSeg/blob/master/tutorials/tutorial-chromosome-1/tutorial-chromosome1_16.md)
- Data for chromosome 1 tutorial [link](https://4dnucleome.cent.uw.edu.pl/PartSeg/Downloads/PartSeg_samples.zip)
- Tutorial: **Different neuron types (as library)** [link](https://github.com/4DNucleome/PartSeg/blob/master/tutorials/tutorial_neuron_types/Neuron_types_example.ipynb)
## Installation
- From binaries:
- [Windows](https://4dnucleome.cent.uw.edu.pl/PartSeg/Downloads/PartSeg-0.14.4-windows.zip) (build on Windows 10)
- [Linux](https://4dnucleome.cent.uw.edu.pl/PartSeg/Downloads/PartSeg-0.14.4-linux.zip) (build on Ubuntu 18.04)
- [MacOS](https://4dnucleome.cent.uw.edu.pl/PartSeg/Downloads/PartSeg-0.14.4-macos.zip) (build on MacOS Mojave)
- With pip:
- From pypi: `pip install PartSeg[all]`
- From repository: `pip install git+https://github.com/4DNucleome/PartSeg.git`
Installation troubleshooting information could be found in wiki: [wiki](https://github.com/4DNucleome/PartSeg/wiki/Instalation-troubleshoot).
If this information does not solve problem you can open [issue](https://github.com/4DNucleome/PartSeg/issues).
## Running
If you downloaded binaries, run the `PartSeg` (or `PartSeg.exe` for Windows) file inside the `PartSeg` folder
If you installed from repository or from pip, you can run it with `PartSeg` command or `python -m PartSeg`.
First option does not work on Windows.
PartSeg export few commandline options:
- `--no_report` - disable error reporting
- `--no_dialog` - disable error reporting and error dialog. Use only when running from terminal.
- `segmentation_analysis` - skip launcher and start analysis gui
- `segmentation` - skip launcher and start segmentation gui
## napari plugin
PartSeg provides napari plugins for io to allow reading projects format in napari viewer.
## Save Format
Saved projects are tar files compressed with gzip or bz2.
Metadata is saved in data.json file (in json format).
Images/masks are saved as *.npy (numpy array format).
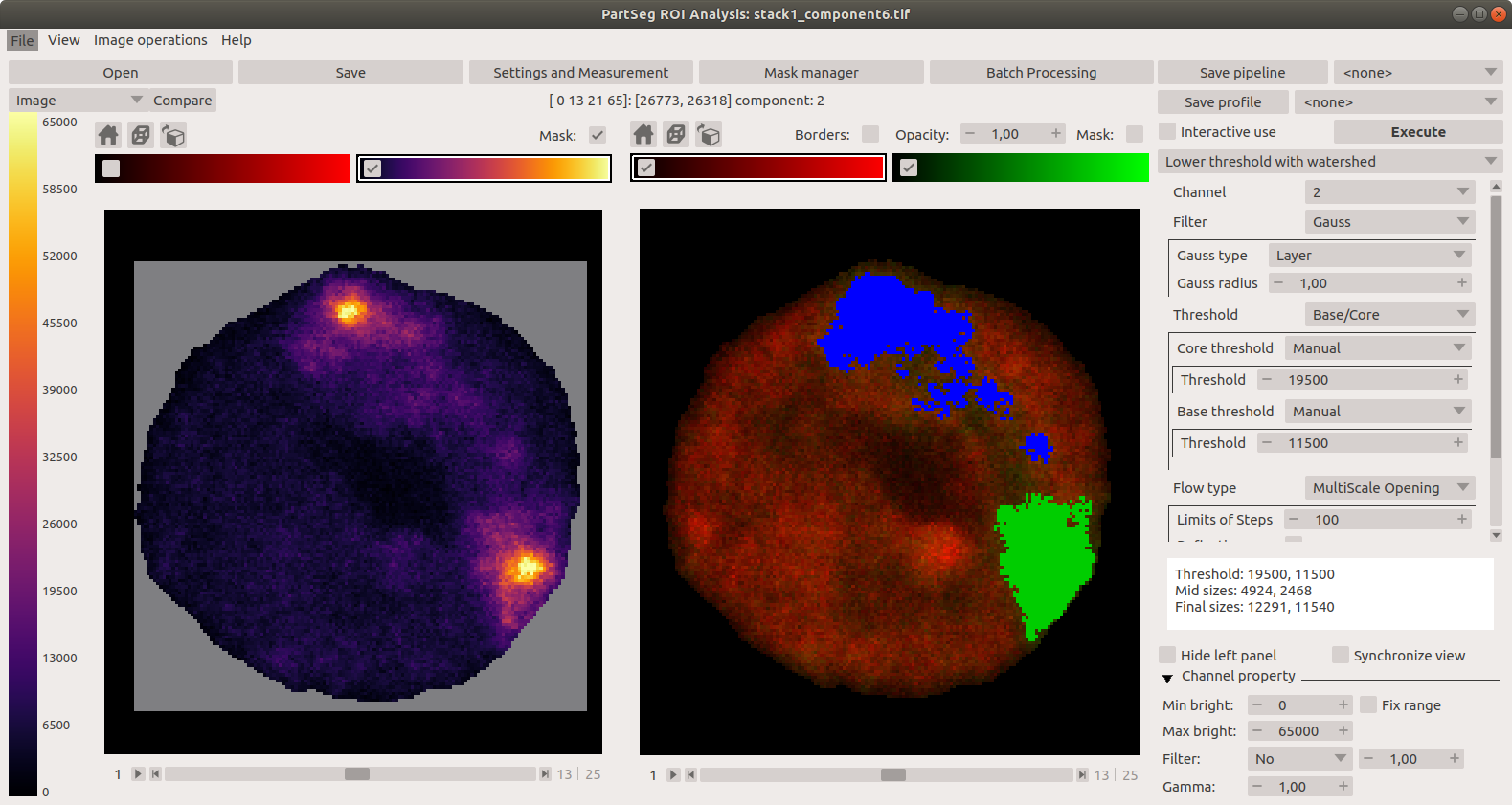
## Interface
Launcher. Choose the program that you will launch:
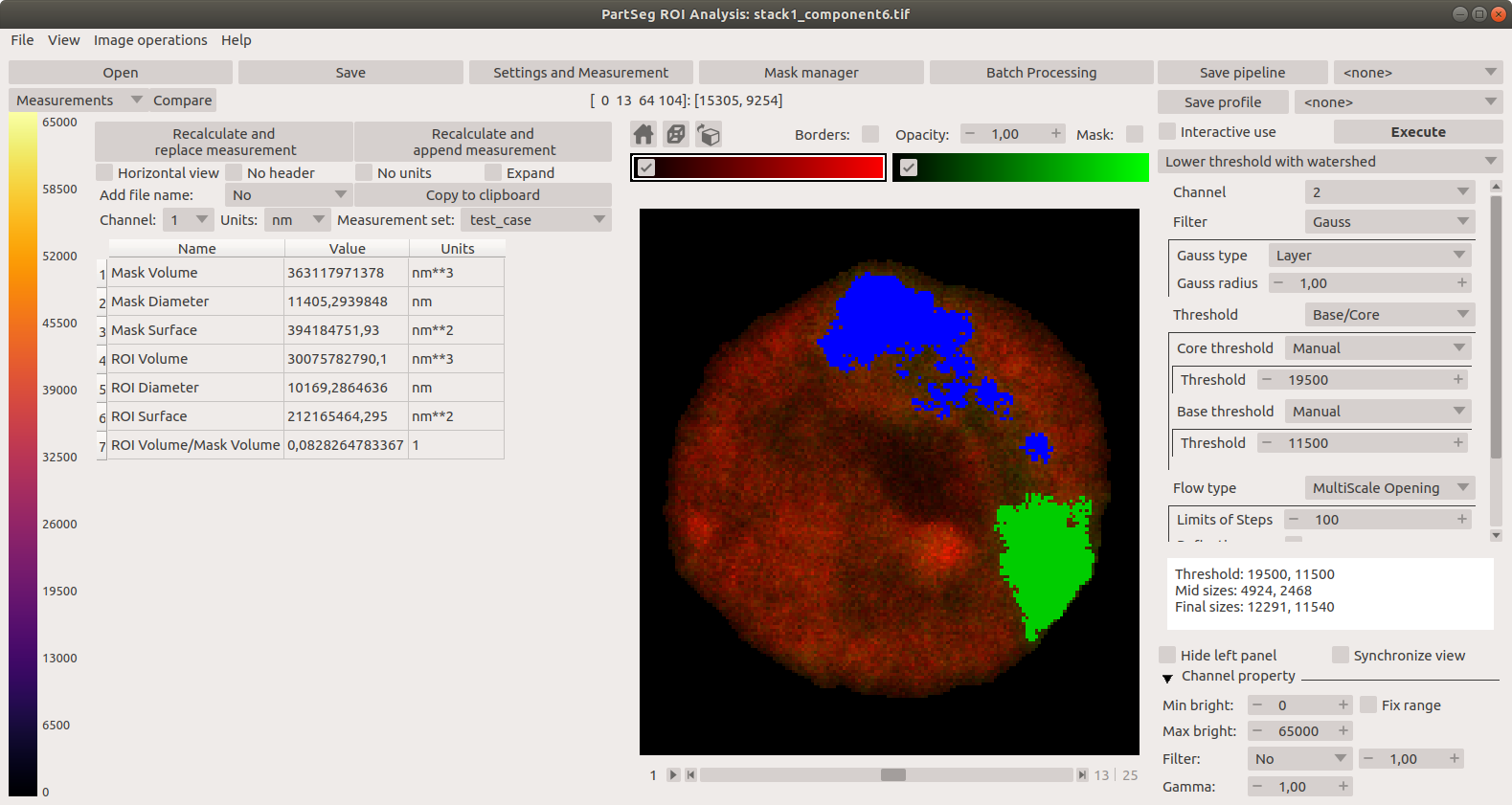
Main window of Segmentation Analysis:

Main window of Segmentation Analysis with view on measurement result:
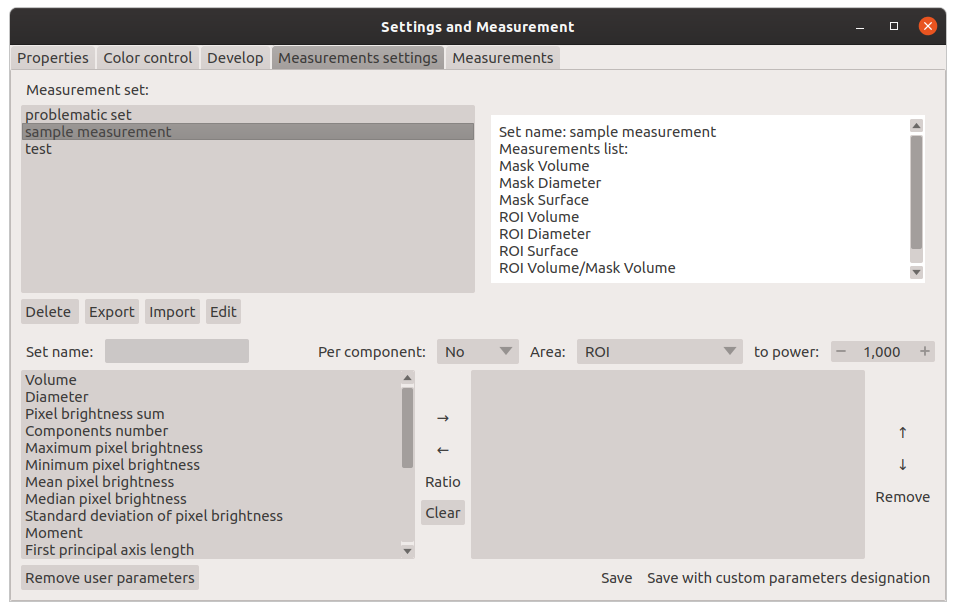
Window for creating a set of measurements:
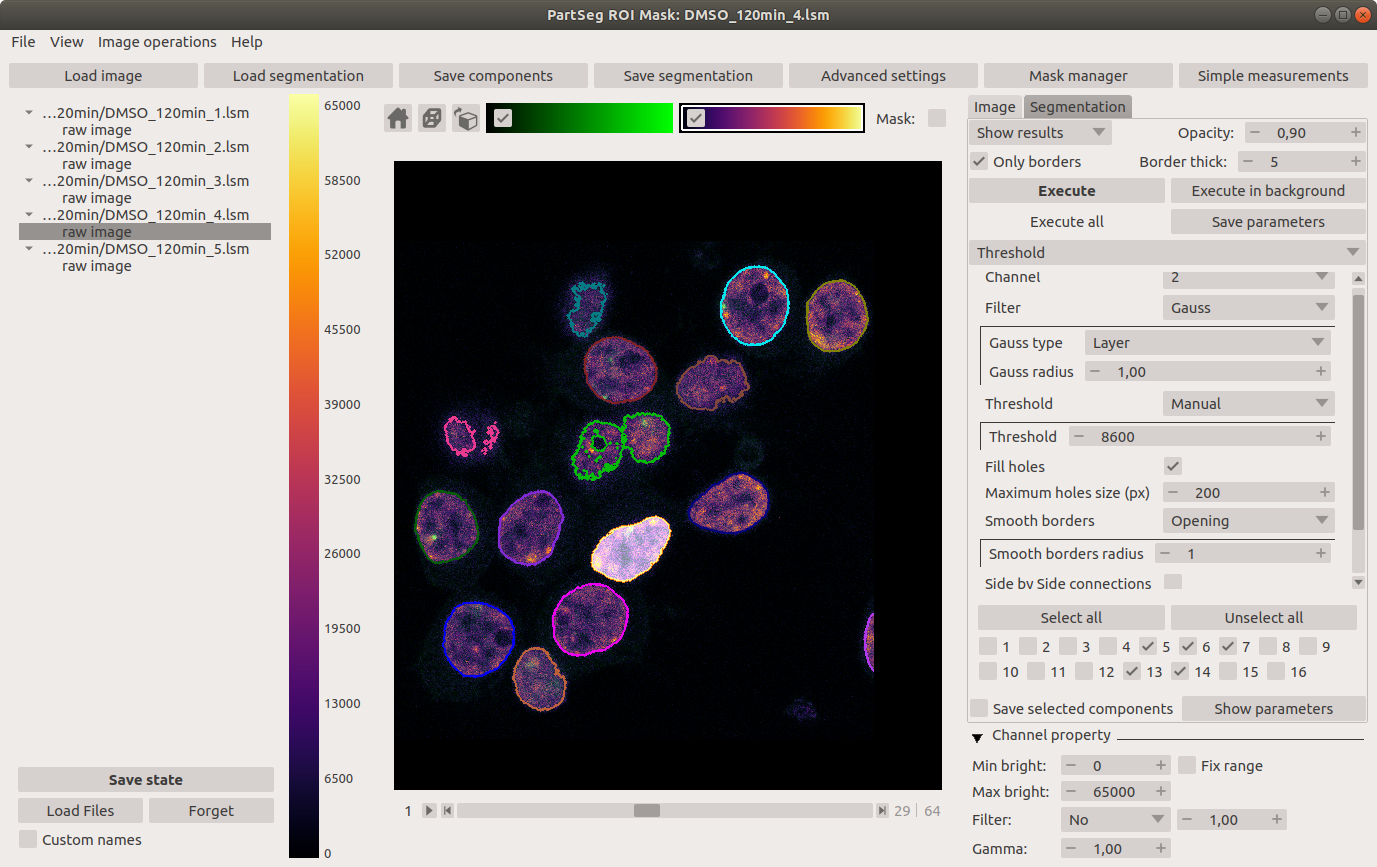
Main window of Mask Segmentation:
## Laboratory
Laboratory of Functional and Structural Genomics
[http://4dnucleome.cent.uw.edu.pl/](http://4dnucleome.cent.uw.edu.pl/)
## Cite as
Bokota, G., Sroka, J., Basu, S. et al. PartSeg: a tool for quantitative feature extraction
from 3D microscopy images for dummies. BMC Bioinformatics 22, 72 (2021).
[https://doi.org/10.1186/s12859-021-03984-1](https://doi.org/10.1186/s12859-021-03984-1)
## Changelog
### 0.14.4 - 2022-10-24
#### Bug Fixes
- Fix `get_theme` calls to prepare for napari 0.4.17 (#729)
- Fix sentry tests (#742)
- Fix reporting error in load settings from the drive (#725)
- Fix saving pipeline from GUI (#756)
- Fix profile export/import dialogs (#761)
- Enable the "Compare" button if ROI is available (#765)
- Fix bug in cut with ROI to not make black artifacts (#767)
#### Features
- Load alternatives labeling when opening PartSeg projects in napari (#731)
- Add option to toggle scale bar (#733)
- Allow customizing the settings directory using the `PARTSEG_SETTINGS_DIR` environment variable (#751)
- Separate recent algorithms from general application settings (#752)
- Add multiple otsu as threshold method with selection range of components (#710)
- Add function to load components from Mask Segmentation with a background in ROI Analysis (#768)
#### Miscellaneous Tasks
- Prepare pyinstaller configuration for napari 0.4.17 (#748)
- Add ruff linter (#754)
#### Testing
- Add new build and inspect wheel action (#747)
#### Build
- Bump actions/checkout from 2 to 3 (#716)
- Bump actions/download-artifact from 1 to 3 (#709)
### 0.14.3 - 2022-08-18
#### Bug Fixes
- Fix lack of rendering ROI when load image from segmentation (#694)
- Fix running ROI extraction from napari widget (#695)
- Delay setting image if an algorithm is still running (#627)
- Wrong error report when no component is found in restartable segmentation algorithm. (#633)
- Fix the process of building documentation (#653)
#### Refactor
- Clean potential vulnerabilities (#630)
#### Testing
- Add more tests for common GUI elements (#622)
- Report coverage per package. (#639)
- Update conda environment to not use PyQt5 in test (#646)
- Add tests files to calculate coverage (#655)
### 0.14.2 - 2022-05-05
#### Bug Fixes
- Fix bug in save label colors between sessions (#610)
- Register PartSeg plugins before starting napari widgets. (#611)
- Mouse interaction with components works again after highlight. (#620)
#### Refactor
- Limit test run (#603)
- Filter and solve warnings in tests (#607)
- Use QAbstractSpinBox.AdaptiveDecimalStepType in SpinBox instead of hardcoded bounds (#616)
- Clean and test `PartSeg.common_gui.universal_gui_part` (#617)
#### Testing
- Speedup test by setup cache for pip (#604)
- Setup cache for azure pipelines workflows (#606)
### 0.14.1 - 2022-04-27
#### Bug Fixes
- Update build wheels and sdist to have proper version tag (#583)
- Fix removing the first measurement entry in the napari Measurement widget (#584)
- Fix compatibility bug for conda Pyside2 version (#595)
- Error when synchronization is loaded, and newly loaded image has different dimensionality than currently loaded. (#598)
#### Features
- Use pygments for coloring code in exception window (#591)
- Add option to calculate Measurement per Mask component (#590)
#### Refactor
- Refactor the creation batch plan widgets and add tests for it (#587)
- Drop napari bellow 0.4.12 (#592)
- Update the order of ROI Mask algorithms to be the same as in older PartSeg versions (#600)
### 0.14.0 - 2022-04-14
#### Bug Fixes
- Fix "Show selected" rendering mode in PartSeg ROI Mask (#565)
- Add access by operator `[]` to `pydantic.BaseModel` base structures for keeping backward compatibility (#579)
#### Features
- Allow setting zoom factor from the interface in Search Label napari plugin (#538)
- Add controlling of zoom factor of search ROI in main GUI (#540)
- Better serialization mechanism allow for declaration data structure migration locally (#462)
- Make `*.obsep" file possible to load in PartSeg Analysis (#564)
- Add option to extract measurement profile or ROI extraction profile from the batch plan (#568)
- Allow import calculation plan from batch result excel file (#567)
- Improve error reporting when failing to deserialize data (#574)
- Launch PartSeg GUI from napari #581
#### Refactor
- Store PartSegImage.Image channels as separated arrays (#554)
- Remove deprecated modules. (#429)
- Switch serialization backend to `nme` (#569)
#### Testing
- Add test of creating AboutDialog (#539)
- Setup test for python 3.10. Disable `class_generator` test for this python (#570)
### 0.13.15
#### Bug Fixes
- Using `translation` instead of `translation_grid` for shifting layers. (#474)
- Bugs in napari plugins (#478)
- Missing mask when using roi extraction from napari (#479)
- Fix segmentation fault on macos machines (#487)
- Fixes for napari 0.4.13 (#506)
#### Documentation
- Create 0.13.15 release (#511)
- Add categories and preview page workflow for the napari hub (#489)
#### Features
- Assign properties to mask layer in napari measurement widget (#480)
#### Build
- Bump qtpy from 1.11.3 to 2.0.0 in /requirements (#498)
- Bump pydantic from 1.8.2 to 1.9.0 in /requirements (#496)
- Bump sentry-sdk from 1.5.1 to 1.5.2 in /requirements (#497)
- Bump sphinx from 4.3.1 to 4.3.2 in /requirements (#500)
- Bump pyinstaller from 4.7 to 4.8 in /requirements (#502)
- Bump pillow from 8.4.0 to 9.0.0 in /requirements (#501)
- Bump requests from 2.26.0 to 2.27.1 in /requirements (#495)
- Bump numpy from 1.21.4 to 1.22.0 in /requirements (#499)
- Bump numpy from 1.22.0 to 1.22.1 in /requirements (#509)
- Bump sphinx from 4.3.2 to 4.4.0 in /requirements (#510)
### 0.13.14
#### Bug Fixes
- ROI alternative representation (#471)
- Change additive to translucent in rendering ROI and Mask (#472)
#### Features
- Add morphological watershed segmentation (#469)
- Add Bilateral image filter (#470)
### 0.13.13
#### Bug Fixes
- Fix bugs in the generation process of the changelog for release. (#428)
- Restoring ROI on home button click in compare viewer (#443)
- Fix Measurement name prefix in bundled PartSeg. (#458)
- Napari widgets registration in pyinstaller bundle (#465)
- Hide points button if no points are loaded, hide Mask checkbox if no mask is set (#463)
- Replace Label data instead of adding/removing layers - fix blending layers (#464)
#### Features
- Add threshold information in layer annotation in the Multiple Otsu ROI extraction method (#430)
- Add option to select rendering method for ROI (#431)
- Add callback mechanism to ProfileDict, live update of ROI render parameters (#432)
- Move the info bar on the bottom of the viewer (#442)
- Add options to load recent files in multiple files widget (#444)
- Add ROI annotations as properties to napari labels layer created by ROI Extraction widgets (#445)
- Add signals to ProfileDict, remove redundant synchronization mechanisms (#449)
- Allow ignoring updates for 21 days (#453)
- Save all components if no components selected in mask segmentation (#456)
- Add modal dialog for search ROI components (#459)
- Add full measurement support as napari widget (#460)
- Add search labels as napari widget (#467)
#### Refactor
- Export common code for load/save dialog to one place (#437)
- Change most of call QFileDialog to more generic code (#440)
#### Testing
- Add test for `PartSeg.common_backend` module (#433)
### 0.13.12
#### Bug Fixes
- Importing the previous version of settings (#406)
- Cutting without masking data (#407)
- Save in subdirectory in batch plan (#414)
- Loading plugins for batch processing (#423)
#### Features
- Add randomization option for correlation calculation (#421)
- Add Imagej TIFF writter for image. (#405)
- Mask create widget for napari (#395)
- In napari roi extraction method show information from roi extraction method (#408)
- Add `*[0-9].tif` button in batch processing window (#412)
- Better label representation in 3d view (#418)
#### Refactor
- Use Font Awesome instead of custom symbols (#424)
### 0.13.11
#### Bug Fixes
- Adding mask in Prepare Plan for batch (#383)
- Set proper completion mode in SearchComboBox (#384)
- Showing warnings on the error with ROI load (#385)
#### Features
- Add CellFromNucleusFlow "Cell from nucleus flow" cell segmentation method (#367)
- When cutting components in PartSeg ROI mask allow not masking outer data (#379)
- Theme selection in GUI (#381)
- Allow return points from ROI extraction algorithm (#382)
- Add measurement to get ROI annotation by name. (#386)
- PartSeg ROI extraction algorithms as napari plugins (#387)
- Add Pearson, Mander's, Intensity, Spearman colocalization measurements (#392)
- Separate standalone napari settings from PartSeg embedded napari settings (#397)
#### Performance
- Use faster calc bound function (#375)
#### Refactor
- Remove CustomApplication (#389)
### 0.13.10
- change tiff save backend to ome-tiff
- add `DistanceROIROI` and `ROINeighbourhoodROI` measurements
### 0.13.9
- annotation show bugfix
### 0.13.8
- napari deprecation fixes
- speedup simple measurement
- bundle plugins initial support
### 0.13.7
- add measurements widget for napari
- fix bug in pipeline usage
### 0.13.6
- Hotfix release
- Prepare for a new napari version
### 0.13.5
- Small fixes for error reporting
- Fix mask segmentation
### 0.13.4
- Bugfix for outdated profile/pipeline preview
### 0.13.3
- Fix saving roi_info in multiple files and history
### 0.13.2
- Fix showing label in select label tab
### 0.13.1
- Add Haralick measurements
- Add obsep file support
### 0.13.0
- Add possibility of custom input widgets for algorithms
- Switch to napari Colormaps instead of custom one
- Add points visualization
- Synchronization widget for builtin (View menu) napari viewer
- Drop Python 3.6
### 0.12.7
- Fixes for napari 0.4.6
### 0.12.6
- Fix prev_mask_get
- Fix cache mechanism on mask change
- Update PyInstaller build
### 0.12.5
- Fix bug in pipeline execute
### 0.12.4
- Fix ROI Mask windows related build (signal not properly connected)
### 0.12.3
- Fix ROI Mask
### 0.12.2
- Fix windows bundle
### 0.12.1
- History of last opened files
- Add ROI annotation and ROI alternatives
- Minor bugfix
### 0.12.0
- Toggle multiple files widget in View menu
- Toggle Left panel in ROI Analysis in View Menu
- Rename Mask Segmentation to ROI Mask
- Add documentation for interface
- Add Batch processing tutorial
- Add information about errors to batch processing output file
- Load image from the batch prepare window
- Add search option in part of list and combo boxes
- Add drag and drop mechanism to load list of files to batch window.
### 0.11.5
- add side view to viewer
- fix horizontal view for Measurements result table
### 0.11.4
- bump to napari 0.3.8 in bundle
- fix bug with not presented segmentation loaded from project
- add frame (1 pix) to image cat from base one based on segmentation
- pin to Qt version to 5.14
### 0.11.3
- prepare for napari 0.3.7
- split napari io plugin on multiple part
- better reporting for numpy array via sentry
- fix setting color for mask marking
### 0.11.2
- Speedup image set in viewer using async calls
- Fix bug in long name of sheet with parameters
### 0.11.1
- Add screenshot option in View menu
- Add Voxels measurements
### 0.11.0
- Make sprawl algorithm name shorter
- Unify capitalisation of measurement names
- Add simple measurements to mask segmentation
- Use napari as viewer
- Add possibility to preview additional output of algorithms (In View menu)
- Update names of available Algorithm and Measurement to be more descriptive.
### 0.10.8
- fix synchronisation between viewers in Segmentation Analysis
- fix batch crash on error during batch run, add information about file on which calculation fails
- add changelog preview in Help > About
### 0.10.7
- in measurements, on empty list of components mean will return 0
### 0.10.6
- fix border rim preview
- fix problem with size of image preview
- zoom with scroll and moving if rectangle zoom is not marked
### 0.10.5
- make PartSeg PEP517 compatible.
- fix multiple files widget on Windows (path normalisation)
### 0.10.4
- fix slow zoom
### 0.10.3
- deterministic order of elements in batch processing.
### 0.10.2
- bugfixes
### 0.10.1
- bugfixes
### 0.10.0
- Add creating custom label coloring.
- Change execs interpreter to python 3.7.
- Add masking operation in Segmentation Mask.
- Change license to BSD.
- Allow select root type in batch processing.
- Add median filter in preview.
### 0.9.7
- fix bug in compare mask
### 0.9.6
- fix bug in loading project with mask
- upgrade PyInstaller version (bug GHSA-7fcj-pq9j-wh2r)
### 0.9.5
- fix bug in loading project in "Segmentation analysis"
### 0.9.4
- read mask segmentation projects
- choose source type in batch
- add initial support to OIF and CZI file format
- extract utils to PartSegCore module
- add automated tests of example notebook
- reversed mask
- load segmentation parameters in mask segmentation
- allow use sprawl in segmentation tool
- add radial split of mask for measurement
- add all measurement results in batch, per component sheet
### 0.9.3
- start automated build documentation
- change color map backend and allow for user to create custom color map.
- segmentation compare
- update test engines
- support of PySide2
### 0.9.2.3
- refactor code to make easier create plugin for mask segmentation
- create class base updater for update outdated algorithm description
- fix save functions
- fix different bugs
### 0.9.2.2
- extract static data to separated package
- update marker of fix range and add mark of gauss in channel control
### 0.9.2.1
- add VoteSmooth and add choosing of smooth algorithm
### 0.9.2
- add pypi base check for update
- remove resetting image state when change state in same image
- in stack segmentation add options to picking components from segmentation's
- in mask segmentation add:
- preview of segmentation parameters per component,
- save segmentation parameters in save file
- new implementation of batch mode.
### 0.9.1
- Add multiple files widget
- Add Calculating distances between segmented object and mask
- Batch processing plan fixes:
- Fix adding pipelines to plan
- Redesign mask widget
- modify measurement backend to allow calculate multi channel measurements.
### 0.9
Begin of changelog




