# Fast Gaussian Accumulator
[](https://pypi.org/project/fastgac/)
[](https://jeremybyu.github.io/FastGaussianAccumulator/)
[](https://github.com/JeremyBYU/FastGaussianAccumulator/actions/workflows/tests.yml)
[](https://github.com/JeremyBYU/FastGaussianAccumulator/blob/master/LICENSE)
A Gaussian Sphere Accumulator refers to the notion of discretizing the **surface** of the unit sphere (a gaussian surface) into buckets/cells. One can then integrate/accumulate a list of **points** (aka unit normals) into these buckets.
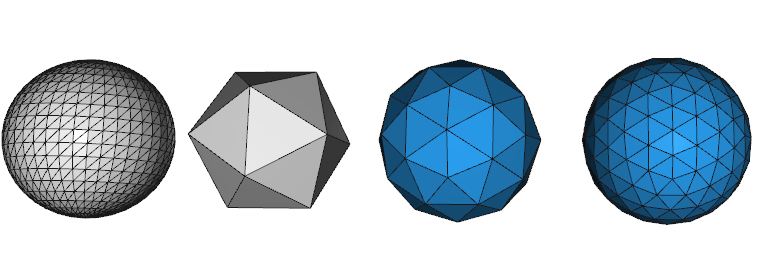
The end result is then a histogram of the sphere. There are many choices for the discretization process, however this library uses equilateral triangles because each cell will have nearly the same **area** and **shape**. This process is done by *refining* an icosahedron. The following image shows our discretization strategy. The first object discretizes a sphere with uniform spacing of phi/theta (note small cells at poles), the second object is an icosahedron, the third object is the first level of refinement for an icosahedron, the last object is the second level of refinement of an icosahedron.
Once a level of refinement is chosen, one can then integrate point vectors into the cells/buckets. For example integrating the normals of a level four (4) icosahedron would look like the image below. Bright yellow indicates more counts for the triangle cells. This is basically showing that the floor [0, 0, 1] and walls [0, +/-1, 0] are common.

## Finding the Cell
To do this one must **find** the cell that corresponds to the point. This is a search process that has been implemented in several fashions in this repo. The main ways are as follows:
* 3D KD Tree - Do a nearest neighbor search using a binary tree.
- `GaussianAccumulatorKDPY` - One implementation using scipy kdtree.
- `GaussianAccumulatorKD` One implementation uses C++ nanoflann.
* Global Index and Local Search - A 3D point is transformed to a unique integer id. The unique ids have the property that ids close to each other will be close to each other in 3D space. The closest id is found corresponding to a triangle cell. A local search of triangle neighbors is performed to find closest triangle cell to the point.
- `GaussianAccumulatorOpt` - Works good on **only** on the top hemisphere. Projects 3D point to plane using Azimuth Equal Area projection. Convert 2D point to int32 index using Hilbert Curve. This implementation is severely limited and is not recommended.
- `GaussianAccumulatorS2Beta` - Works on full sphere! Uses Googles S2 space filling curve (uint64). 3D point is projected to unit cube, assigned to a face of the cube, and then a Hilbert curve index is found for that cube face. This is recommended, and what I use.
Use GaussianAccumulatorS2Beta! Look at `python -m examples.python.run_normals`
## Peak Detection
There are two (2) peak detection methods of used within this repository. The user can choose which one best suite there needs.
### 2D Image Peak Detection
This method basically unwraps the icosahedron as a 2D image in a very particular way as described by [Gauge Equivariant Convolutional Networks and the Icosahedral CNN]("https://arxiv.org/abs/1902.04615). This unwrapping is hardcoded and fixed once a refinement level is chosen so its very fast. The library then uses a 2D peak detector algorithm followed up with agglomerative hierarchial clustering (AHC) to group similar peaks. All of this is user configurable.
### 1D Signal Peak Detection
This performs peak detection on the 1D thread following the hilbert curve. This produces more peaks which are actually near each other on S2 and are then grouped with AHC. This actually works pretty well, but I recommend to use the 2D Image Peak Detector.
## Installation
For python there are pre-built binary wheel on PyPI for Windows and Linux. You can install with `pip install fastgac`.
Below are instruction to build the C++ Package (and python package) manaully with CMake. Installation is entirely through CMake now. You must have CMake 3.14 or higher installed and a C++ compiler with C++ 14 or higher.
### Build Project Library
1. `mkdir cmake-build && cd cmake-build`. - create build folder directory
2. `cmake ../ -DCMAKE_BUILD_TYPE=Release` . For windows also add `-DCMAKE_GENERATOR_PLATFORM=x64`
3. `cmake --build . -j$(nproc) --config Release` - Build FastGA
### Build and Install Python Extension
#### For Developers
1. Install [conda](https://conda.io/projects/conda/en/latest/) or create a python virtual environment ([Why?](https://medium.freecodecamp.org/why-you-need-python-environments-and-how-to-manage-them-with-conda-85f155f4353c)). I recommend conda for Windows users.
2. `cd cmake-build && cmake --build . --target python-package --config Release -j$(nproc)`
3. `cd lib/python_package && pip install -e .`
If you want to run the examples then you need to install the following (from main directory):
1. `pip install -r dev-requirements.txt`
#### For Python Users (Requires CMake)
1. Install [conda](https://conda.io/projects/conda/en/latest/) or create a python virtual environment ([Why?](https://medium.freecodecamp.org/why-you-need-python-environments-and-how-to-manage-them-with-conda-85f155f4353c)). I recommend conda for Windows users.
2. `pip install .`
If you want to run the examples then you need to install the following (from main directory):
1. `pip install -r dev-requirements.txt`
<!-- ### Build with S2Geometry
Googles S2Geometry library is **not** needed in this repository. I have encapsulated **all** code that transforms a unit normal (xyz point) to a unique integer id (uint64) into the header only file `include/NanoS2ID/NanoS2ID.hpp`. However if you desire to run some benchmarks comparing S2 geometry code with this `nano2sid` you must install download and install S2 but first applying the following patch to the source code.
To build with S2 you must apply this patch first.
```
diff --git a/CMakeLists.txt b/CMakeLists.txt
index 5ecd280..d67bf76 100644
--- a/CMakeLists.txt
+++ b/CMakeLists.txt
@@ -411,7 +411,7 @@ install(TARGETS s2 s2testing DESTINATION lib)
message("GTEST_ROOT: ${GTEST_ROOT}")
if (GTEST_ROOT)
* add_subdirectory(${GTEST_ROOT} build_gtest)
+# add_subdirectory(${GTEST_ROOT} build_gtest)
include_directories(${GTEST_ROOT}/include)
set(S2TestFiles
```
Then enable the option for CMake. -->
## Documentation
Please see [documentation website](https://jeremybyu.github.io/FastGaussianAccumulator/) for more details.
<!-- ## General Notes
The main class of interest should be `GaussianAccumulatorS2`
`NanoS2ID.hpp` is header only and which allows it to be optimized more easily (for developer). `S2Geometry` can be compiled as a shared library or a static library (I have tried both). Getting an S2ID is about 50% faster using `NanoS2ID` . My guess is that there is function call overhead in calling a library, vs inlining the function. However, I did build S2 as a static library and did enable link time optimization, but it didn't make it any faster. However I'm guessing I just made a mistake in this process and its *possible* to make `S2Geometry` as fast as `NanoS2ID` .
I also tried using S2 as a point index (similar to a KDTree) and found it was *significantly* slower, about 5X slower than using a KDTree. -->
<!--
### Profiling
1. `LD_PRELOAD=/usr/lib/x86_64-linux-gnu/libprofiler.so CPUPROFILE=prof.prof CPUPROFILE_FREQUENCY=1000`
2. `google-pprof --cum --web ./cmake-build/bin/example-kd prof.prof`
-->
## Citation
To support our work please cite:
```
@Article{s20174819,
AUTHOR = {Castagno, Jeremy and Atkins, Ella},
TITLE = {Polylidar3D-Fast Polygon Extraction from 3D Data},
JOURNAL = {Sensors},
VOLUME = {20},
YEAR = {2020},
NUMBER = {17},
ARTICLE-NUMBER = {4819},
URL = {https://www.mdpi.com/1424-8220/20/17/4819},
ISSN = {1424-8220}
}
```
<!--
```
{
"class_name" : "ViewTrajectory",
"interval" : 29,
"is_loop" : false,
"trajectory" :
[
{
"boundingbox_max" : [ 1.1394986767814996, 1.1386749634896274, 1.1998667377539569 ],
"boundingbox_min" : [ -1.1335375159673458, -1.0, -1.1990006616678834 ],
"field_of_view" : 60.0,
"front" : [ -0.98297198230758687, 0.069800731498603108, 0.16998217518479183 ],
"lookat" : [ 0.0029805804070769382, 0.069337481744813689, 0.00043303804303673754 ],
"up" : [ -0.033111791938401081, 0.84262061564110757, -0.53748870437547691 ],
"zoom" : 0.5199999999999998
}
],
"version_major" : 1,
"version_minor" : 0
}
```
-->




