# decOM: Similarity-based microbial source tracking of ancient oral samples using k-mer-based methods
`decOM` is a high-accuracy microbial source tracking method that is suitable for contamination quantification in paleogenomics, namely the analysis of collections of possibly contaminated ancient oral metagenomic data sets. **In few words**, if you want to know how contaminated your ancient oral metagenomic sample is, this tool will help 🧹🦷🧹
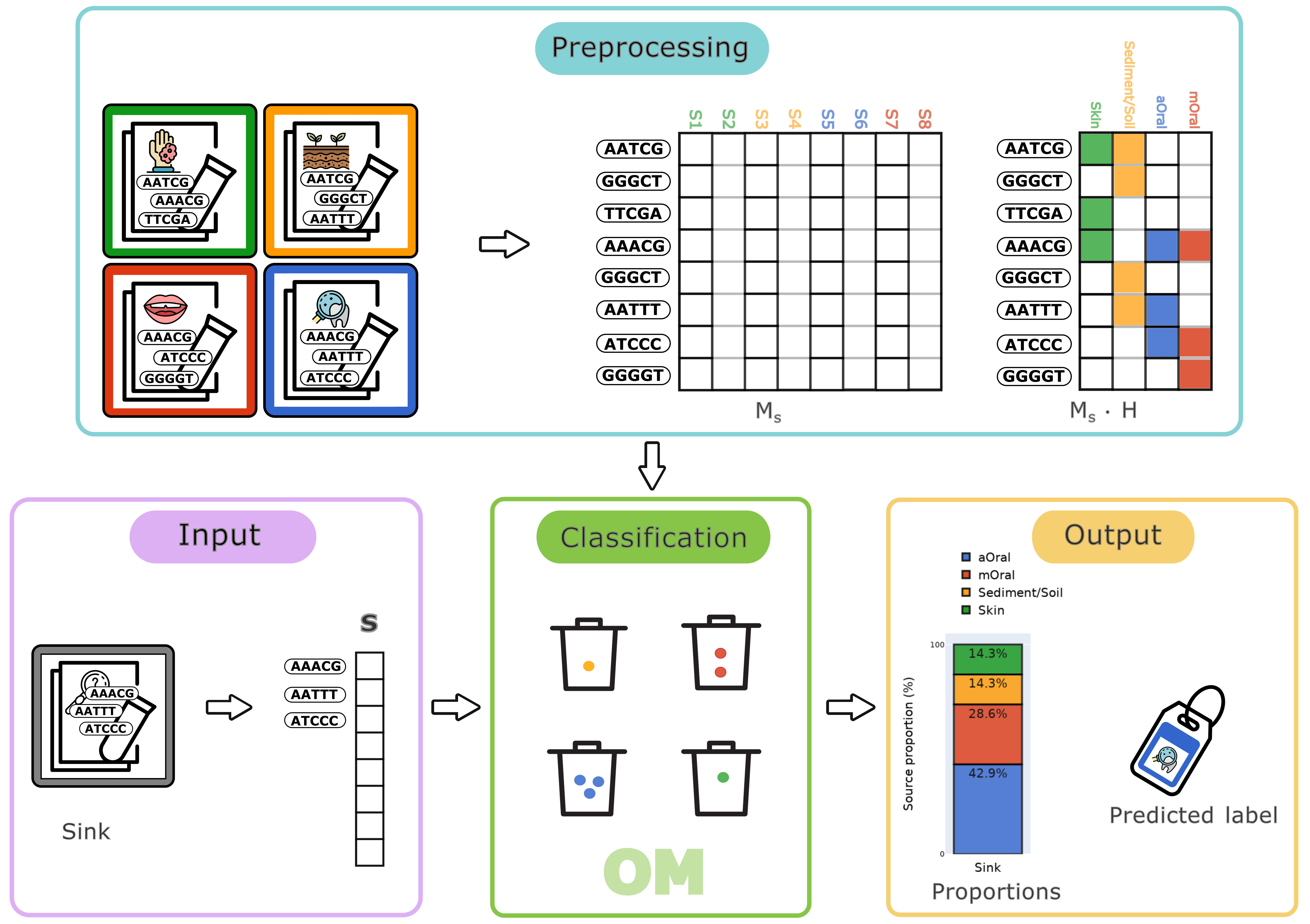
**In longer words** `decOM` comes with a k-mer matrix of ancient Oral(aOral) metagenomic samples and its possible contaminants: Sediment/Soil, Skin and modern oral (mOral). It represents every sink the user inputs as a presence/absence vector of k-mers and then estimates and outputs the proportions of each source environment in the sink. Ex: K-mer AAACG is present in the input sink S and in source S1 labelled as Skin, S5 labelled as aOral and S7 labelled as mOral, hence one ball is added to the bin of Skin, aOral and mOral respectively. After every entry in the the sink vector is compared against every entry of every vector in the sources, decOM outputs the estimated environment proportions and the hard label assigned to the sink $s$ is that of the environment with the highest contribution.
+ [System requirements](#system-requirements)
+ [Installation](#installation)
+ [Before running decOM](#before-running-decom)
+ [Test](#test)
+ [Output files](#output-files)
+ [Usage](#usage)
+ [Additional features](#additional-features)
+ [aOralOut](#aoralout)
+ [MST](#MST)
+ [LOO](#LOO)
+ [Command-line options](#command-line-options)
## System requirements
`decOM` has been developed and tested under a Linux environment and it only works in Linux-like systems.
It requires certain packages/tools in order to be installed/used:
+ [miniconda3](https://conda.io/en/latest/miniconda.html)
## Installation
Install `decOM` through conda:
```
git clone https://github.com/CamilaDuitama/decOM.git
cd decOM
conda env create -n decOM --file environment.yml
conda deactivate
conda activate decOM
```
To make the ``decOM`` command available, it is advised to include the absolute path of `decOM` in your PATH environment variable by adding the following line to your `~/.bashrc` file:
```
export PATH=/absolute/path/to/decOM:${PATH}
```
## Before running decOM
**BEFORE** running `decOM` you must first download the folder [decOM_sources.tar.gz](https://zenodo.org/record/6513520/files/decOM_sources.tar.gz) and decompress it. You can either follow the link or use wget (it has to be installed in your computer first):
```
wget https://zenodo.org/record/6513520/files/decOM_sources.tar.gz
tar -xf decOM_sources.tar.gz
```
The path of the extracted directory `decOM_sources` will be requested through the input parameter `-p_sources` each time `decOM` is run (see examples below).
## Test
### Single sink
You can test if `decOM` is working by using the aOral sample present in the `test/sample/` folder, ex: SRR13355807.
```
decOM -s SRR13355807 -p_sources decOM_sources/ -k tests/sample/SRR13355807.fof -mem 10GB -t 5
```
*Note*: The final memory allocated for each run of `decOM` will be your input in `-mem` times the number of cores (`-t`). In the previous run we used 10GB * 5 = 50 GB. It is recommended to run `decOM` with at least 10GB of memory and 1 core.
### Several sinks
You can test if `decOM` with several sinks by using the files inside `test/several_samples/` as follows:
```
decOM -p_sources decOM_sources/ -p_sinks tests/several_samples/sinks.txt -p_keys tests/several_samples/ -mem 10GB -t 5
```
`decOM` relies on `DASK` for parallelization. Once you start running `decOM` and the client is set, you can see the [diagnostic dashboard](https://docs.dask.org/en/stable/dashboard.html) to follow the process and better tune parameters such as `-mem `and `-t`, make sure you can connect to your local host and visualise it here: http://127.0.0.1:8787/status
## Output files
`decOM` will output one .csv file with the k-mer counts and proportions, a folder with the vector representing the sample of interest, from now on called sink (s), and a barplot if indicated by the user.
```
decOM_output/
├──decOM_output.csv
├──result_plot_sinks.pdf
├──result_plot_sinks.html
├──{s}_vector/
```
The `decOM_output.csv` file is a dataframe that contains one row per sink. The columns correspond to the raw number of k-mers per source environment,
the running time per sink, the sink name and the proportions. The result for the one sample explained before should look like this:
| Sediment/Soil | Skin | aOral | mOral | Unknown | Running time (s) | Sink | p_Sediment/Soil | p_Skin | p_aOral | p_mOral | p_Unknown | decOM_max |
| :----: | :----: | :----: | :----: | :----: |:----------------:| :----: | :----: | :----: | :----: | :----: | :----: | :----: |
| 182 | 281 | 197859 | 37023 | 334 |196.7268| SRR13355807 | 0.0772 | 0.1192 | 83.9527 | 15.7091 | 0.1417 | aOral |
The `result_plot_sinks.pdf` and `result_plot_sinks.html` are static and interactive plots (respectively) for the proportions of source environments per sink.
The `{s}_vector/` folder is the output of kmtricks filter + kmtricks aggregate.
> **UNKOWNS IS A FEATURE IN DEVELOPMENT!**: The contribution of an unknown source is a feature in development. For a more accurate assessment of the contamination proportions in a sample, please take into account the raw counts for Sediment/Soil, Skin, aOral, mOral.
## Usage
You can use as input your fastq/fasta file from your own experiment, you can download an ancient oral sample of interest from the [AncientMetagenomeDir](https://github.com/SPAAM-community/AncientMetagenomeDir) or from the [SRA](https://sra-explorer.info/).
The users of `decOM` can represent their own metagenomic sample as a presence/absence vector of k-mers using kmtricks. This sink can be compared against the collection of sources we have put together.
Once you have downloaded the folder with the [matrix of sources](#before-running-decom) and the fastq file(s) of your sink(s), you have to create a `key.fof` file per sink.
The `key.fof` has one line of text depending on your type of data:
**-Single-end:**
`s : path/to/file/s_1.fastq.gz`
**-Paired-end :**
`s : path/to/file/s_1.fastq.gz; path/to/file/s_2.fastq.gz `
*Note*: As `decOM` relies on [`kmtricks`](https://github.com/tlemane/kmtricks), you might use a FASTA or FASTQ format, gzipped or not, which means you have to change the `key.fof` file accordingly.
Since you now have the fasta/fastq file of your sink, the folder with the matrix of sources and the key file, simply run `decOM` as follows:
### Single sink
```decOM -s {SINK} -p_sources decOM_sources/ -k {KEY.FOF} -mem {MEMORY} -t {THREADS}```
The parameter `-s` refers to the name of the sink, whereas the parameter `-k` is the path to the **file** `key.fof`. Make sure that the name you use for your sink is the same as the name you use in the `key.fof`. So if your sink is called s, `-s s_1`, your `key.fof` should be `s_1 : s_1.fastq`
### Several sinks
If you want to assess the contamination of several sinks, you need one `key.fof` file per sink, and they must be inside the folder `p_sources`
```decOM -p_sinks {PATH_SINKS} -p_sources decOM_sources/ -p_keys {PATH_KEYS} -mem {MEMORY} -t {THREADS}```
The parameter `-p_sinks` refers to a .txt **file** with a list of sinks limited by a newline (\n). Each sink in this file must have a corresponding `key.fof` file in the folder `p_keys/` . While `p_sinks` is the path to a **file**, `p_keys` is a path to a **folder** that should contain as many `key.fof` files as sinks.
If you have *n* samples, your the content of the file `p_sinks` should be:
```
s_1
s_2
s_3
.
.
.
s_n
```
And the content of `p_keys` should be:
```
p_keys/
├──s_1.fof
├──s_2.fof
├──s_3.fof
.
.
.
├──s_n.fof
```
## Additional features
### aOralOut:
This feature was thought for the users who prefered having mOral, Skin and Sediment/Soil samples as sources but no aOral samples. Once you download and unzip the new matrix of sources:
```
wget https://zenodo.org/record/6772124/files/aOralOut_sources.tar.gz
tar -xf aOralOut_sources.tar.gz
```
Simply run `decOM-aOralOut` as follows:
````
decOM-aOralOut -s SRR13355807 -p_sources aOralOut_sources/ -k tests/sample/SRR13355807.fof -mem 10GB -t 5
````
### MST:
This feature was thought for the users who prefer building their own k-mer matrix of sources. To run `decOM-MST` you need to create your own `p_sources` folder and additionally you need a `-m` or map file with one label per source.
To create the `p_sources` you can run [kmtricks](https://github.com/tlemane/kmtricks/wiki) (already in your conda environment for decOM) as follows:
```
kmtricks pipeline --file kmtricks.fof --run-dir p_sources --mode kmer:pa:bin
kmtricks aggregate --run-dir p_sources --pa-matrix kmer --output p_sources/matrices/matrix.pa --format bin
kmtricks dump --run-dir p_sources --input p_sources/matrices/matrix.pa.lz4 -o p_sources/matrices/matrix.pa.txt
```
**NOTE:** Building a k-mer matrix with any other parameters of kmtricks and using it as input for `decOM-MST` has not been tested.
You additionally need a `-m` file which is a .csv file of two columns: *Env* and *SampleID*. This is a `x` by 2 table, where `x` is the number of sources in your input k-mer matrix (number of columns in the kmtricks.fof used to run kmtricks). `SampleID` refers to the unique identifier of each source sample, and `Env` is the corresponding label for the source environment from where each sample was taken.
You can run `decOM-MST` with test data as follows:
```
kmtricks pipeline --file tests/MST/kmtricks.fof --run-dir p_sources --mode kmer:pa:bin
kmtricks aggregate --run-dir p_sources --pa-matrix kmer --output p_sources/matrices/matrix.pa --format bin
kmtricks dump --run-dir p_sources/ --input p_sources/matrices/matrix.pa -o p_sources/matrices/matrix.pa.txt
decOM-MST -s SRR13355807 -p_sources p_sources/ -m tests/MST/map.csv -k tests/sample/SRR13355807.fof --mem 10GB -t 5
```
### LOO:
This feature was thought for the users who would like to perform a leave-one-out experiment on their own matrix of sources. This experiment consist on creating a k-mer matrix of sources, extracting one sample to be used as sink and leave the reas out as sources. Once this iteration is finished, `decOM-LOO` would take out a different sink from the matrix of sources, and compare it against the rest of samples.`decOM-LOO` stops once every sample has been compared against the rest. To run `decOM-LOO` you need to create your own `p_sources` folder and additionally you need a `-m` or map file with one label per source. **NOTE**:This feature will only work if there are at least 2 samples representative of each source environment
To create the `p_sources` you can run [kmtricks](https://github.com/tlemane/kmtricks/wiki) (already in your conda environment for decOM) as follows:
```
kmtricks pipeline --file kmtricks.fof --run-dir p_sources --mode kmer:pa:bin
kmtricks aggregate --run-dir p_sources --pa-matrix kmer --output p_sources/matrices/matrix.pa --format bin
kmtricks dump --run-dir p_sources --input p_sources/matrices/matrix.pa.lz4 -o p_sources/matrices/matrix.pa.txt
```
**NOTE:** Building a k-mer matrix with any other parameters of kmtricks and using it as input for `decOM-LOO` has not been tested.
You additionally need a `-m` file which is a .csv file of two columns: *Env* and *SampleID*. This is a `x` by 2 table, where `x` is the number of sources in your input k-mer matrix (number of columns in the kmtricks.fof used to run kmtricks). `SampleID` refers to the unique identifier of each source sample, and `Env` is the corresponding label for the source environment from where each sample was taken.
You can run `decOM-LOO` with test data as follows:
```
kmtricks pipeline --file tests/LOO/kmtricks.fof --run-dir p_sources --mode kmer:pa:bin
kmtricks aggregate --run-dir p_sources --pa-matrix kmer --output p_sources/matrices/matrix.pa --format bin
kmtricks dump --run-dir p_sources/ --input p_sources/matrices/matrix.pa -o p_sources/matrices/matrix.pa.txt
decOM-LOO -p_sources p_sources/ -m tests/LOO/map.csv --mem 10GB -t 5
```
## Command line options
```
usage: decOM [-h] (-s SINK | -p_sinks PATH_SINKS) -p_sources PATH_SOURCES (-k KEY | -p_keys PATH_KEYS) -mem MEMORY -t THREADS [-o OUTPUT] [-p {True,False}] [-V] [-v]
Microbial source tracking for contamination assessment of ancient oral samples using k-mer-based methods
Argurguments:
-h, --help show this help message and exit
-s SINK, --sink SINK Write down the name of your sink. It must be the same as the first element of key.fof. When this argument is set, -k/--key must be defined too
-p_sinks PATH_SINKS, --path_sinks PATH_SINKS
.txt file with a list of sinks limited by a newline (\n). When this argument is set, -p_keys/--path_keys must be defined too.
-p_sources PATH_SOURCES, --path_sources PATH_SOURCES
path to folder downloaded from https://zenodo.org/record/6513520/files/decOM_sources.tar.gz
-k KEY, --key KEY filtering key (a kmtricks fof with only one sample). When this argument is set, -s/--sink must be defined too.
-p_keys PATH_KEYS, --path_keys PATH_KEYS
Path to folder with filtering keys (a kmtricks fof with only one sample).You should have as many .fof files as sinks.When this argument is set,
-p_sinks/--path_sinks must be defined too.
-mem MEMORY, --memory MEMORY
Write down how much memory you want to use for this process. Ex: 10GB
-t THREADS, --threads THREADS
Number of threads to use. Ex: 5
-o OUTPUT, --output OUTPUT
Path to output folder, where you want decOM to write the results. Folder must not exist, it won't be overwritten.
-p {True,False}, --plot {True,False}
True if you want a plot (in pdf and html format) with the source proportions of the sink, else False
-V, --version Show version number and exit
-v, --verbose Verbose output
```




