# Dash JBrowse
[Dash](https://dash.plotly.com/introduction) is a popular open-source framework for building data-driven web apps.
It is built on top of ReactJS, and provides a [toolkit](https://dash.plotly.com/plugins) for converting React components into dash components that can be used in Dash apps written in Python, R, Julia, F# or Matlab.
Dash JBrowse's `LinearGenomeView` is a Dash component that wraps the [JBrowse React Linear Genome View](https://jbrowse.org/storybook/lgv/main/) and makes it possible to embed an interactive genome browser into any Dash application. Now supporting the `CircularGenomeView` as well.
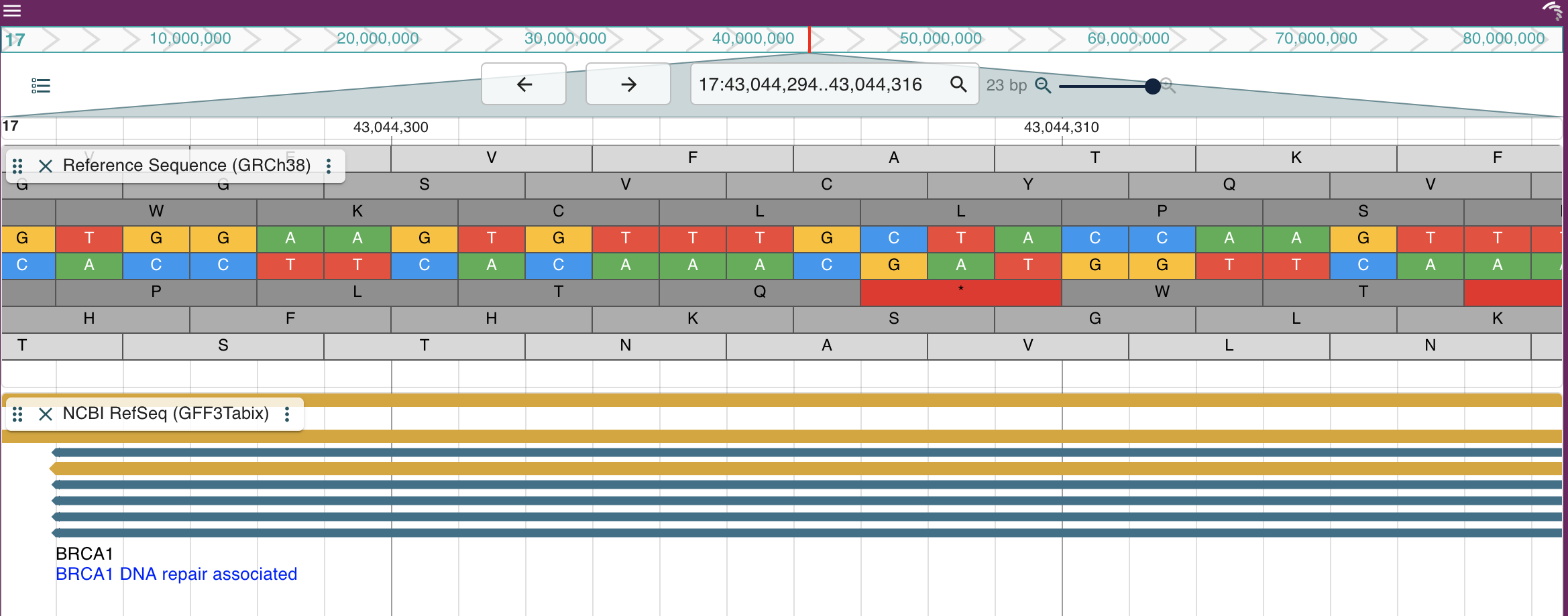
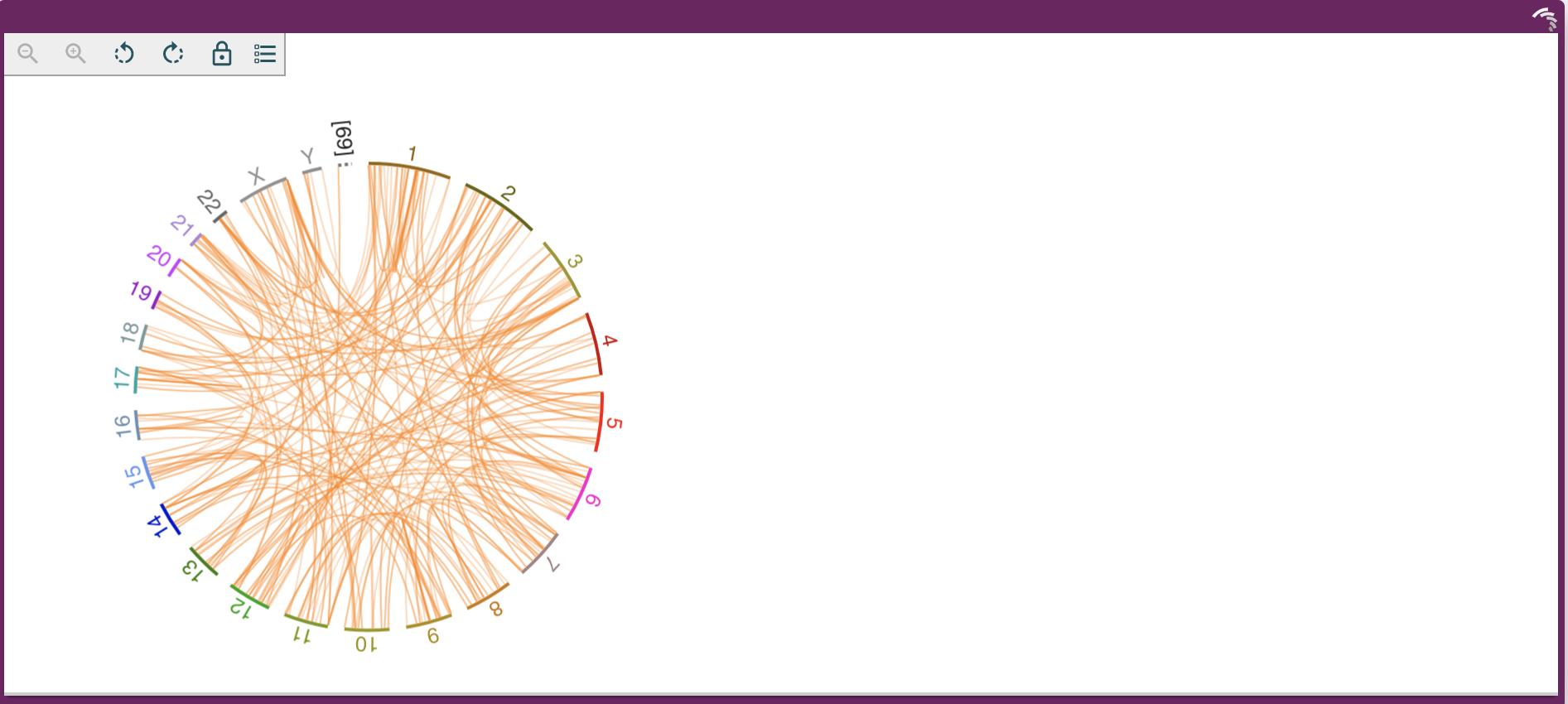
## Quickstart
```
$ pip install dash_jbrowse
```
Basic linear genome view with an assembly
```
import dash
import dash_jbrowse
from dash import html
print(dash_jbrowse.__version__)
app = dash.Dash(__name__)
my_assembly = {
"name": "GRCh38",
"sequence": {
"type": "ReferenceSequenceTrack",
"trackId": "GRCh38-ReferenceSequenceTrack",
"adapter": {
"type": "BgzipFastaAdapter",
"fastaLocation": {
"uri": "https://s3.amazonaws.com/jbrowse.org/genomes/GRCh38/fasta/GRCh38.fa.gz",
},
"faiLocation": {
"uri": "https://s3.amazonaws.com/jbrowse.org/genomes/GRCh38/fasta/GRCh38.fa.gz.fai",
},
"gziLocation": {
"uri": "https://s3.amazonaws.com/jbrowse.org/genomes/GRCh38/fasta/GRCh38.fa.gz.gzi",
},
},
},
"aliases": ["hg38"],
"refNameAliases": {
"adapter": {
"type": "RefNameAliasAdapter",
"location": {
"uri": "https://s3.amazonaws.com/jbrowse.org/genomes/GRCh38/hg38_aliases.txt",
},
},
},
}
app.layout = html.Div(
[
dash_jbrowse.LinearGenomeView(
id="lgv-hg38",
assembly=my_assembly
),
],
id='test'
)
if __name__ == "__main__":
app.run_server(debug=True)
```
An example of the CGV can be found in [usage_cgv.py](usage_cgv.py).
## Installation
### PyPI
```
$ pip install dash_jbrowse
```
### Install with virtual venv
After cloning this repository, install virtualenv
```
$ pip install virtualenv
```
```
$ cd dash_jbrowse
$ python3 -m venv ./venv
$ source venv/bin/activate
$ pip install -r requirements.txt
```
Then you can run the example app found in usage.py
```
python usage.py
```
## User Guide
The views can be customized in many ways, but the only properties that are required to launch it are the id required for dash call backs and the assembly.
- `id` (string, required): the id used for dash callbacks.
A basic example of this component using dash callbacks can be found under the examples directory)
- `assembly` (object, required): the configuration object of the assembly being used for the browser. More information about the configuration of the assembly can be found [here](https://jbrowse.org/jb2/docs/config_guide/#configuring-assemblies).
_top level assembly config_
```json
{
"name": "assemblyName",
"aliases": [],
"sequence": {},
"refNameAliases": {}
}
```
- `tracks` (list, optional) - list of track configuration objects.
_top level track config_
```json
{
"trackId": "test-id",
"name": "track name",
"assemblyNames": [],
"category": []
}
```
- `defaultSession` (object, optional) - information about the current app state such as what views are open
_default session config_
```json
{
"name": "this session",
"view": {
"id": "linearGenomeView",
"type": "LinearGenomeView"
}
}
```
Note: `"type": "CircularGenomeView"` for CGV
- `location` (string or object, optional) - initial [location](https://jbrowse.org/jb2/docs/user_guide/#using-the-location-search-box) for when the browser first loads, e.g '1:500-1000' or location object
_location object_
```json
{
"refName": 1,
"start": 500,
"end": 1000
}
```
Note:
* Use 0-based coordinates in the location object.
* The location field is only used for LGV and not CGV.
Checkout the storybook [JBrowse React Linear Genome View docs](https://jbrowse.org/storybook/lgv/main/?path=/story/default-sessions--page) or the [JBrowse React Circular Genome View docs](https://jbrowse.org/storybook/cgv/v1.5.9/?path=/story/getting-started--page) for more configuration examples.
## Advanced Customization
### Text Searching
Adding text searching to the JBrowse React Linear Genome View (not currently available for the CGV) requires a couple of extra steps. We first need to create an index via the [JBrowse CLI tools](https://jbrowse.org/jb2/docs/cli/#jbrowse-text-index). Once an index is created, we can add a text search adapter to the list of aggregate text search adapters in this component or to the configuration object of a track.
- `aggregateTextSearchAdapters` (object, optional) - configuration of an index used for text searching
Aggregate text search adapter to use in the component.
```json
{
"type": "TrixTextSearchAdapter",
"textSearchAdapterId": "adapter-id",
"ixFilePath": {
"uri": "url/path/to/my/ix/file",
"locationType": "UriLocation"
},
"ixxFilePath": {
"uri": "url/path/to/my/ixx/file",
"locationType": "UriLocation"
},
"metaFilePath": {
"uri": "url/path/to/my/meta.json/file",
"locationType": "UriLocation"
}
}
```
On a track configuration.
```json
{
"trackId": "yourtrack",
"name": "Track name",
"adapter": {
"type": "Gff3TabixAdapter",
"gffGzLocation": {
"uri": "yourfile.gff.gz",
"locationType": "UriLocation"
},
"index": {
"location": {
"uri": "yourfile.gff.gz.tbi",
"locationType": "UriLocation"
}
}
},
"textSearching": {
"textSearchAdapter": {
"type": "TrixTextSearchAdapter",
"textSearchAdapterId": "hg19-index",
"ixFilePath": {
"uri": "https://jbrowse.org/genomes/hg19/trix/hg19.ix",
"locationType": "UriLocation"
},
"ixxFilePath": {
"uri": "https://jbrowse.org/genomes/hg19/trix/hg19.ixx",
"locationType": "UriLocation"
},
"metaFilePath": {
"uri": "https://jbrowse.org/genomes/hg19/trix/meta.json",
"locationType": "UriLocation"
},
"assemblyNames": ["hg19"]
},
"indexingAttributes": ["Name", "ID"],
"indexingFeatureTypesToExclude": ["CDS", "exon"]
}
}
```
### Custom themes are also available.
- `configuration` - (object, optional) - color scheme configuration
```json
{
"theme": {
"palette": {
"primary": {
"main": "#311b92"
},
"secondary": {
"main": "#0097a7"
},
"tertiary": {
"main": "#f57c00"
},
"quaternary": {
"main": "#d50000"
},
"bases": {
"A": {"main": "#98FB98"},
"C": {"main": "#87CEEB"},
"G": {"main": "#DAA520"},
"T": {"main": "#DC143C"}
}
}
}
}
```
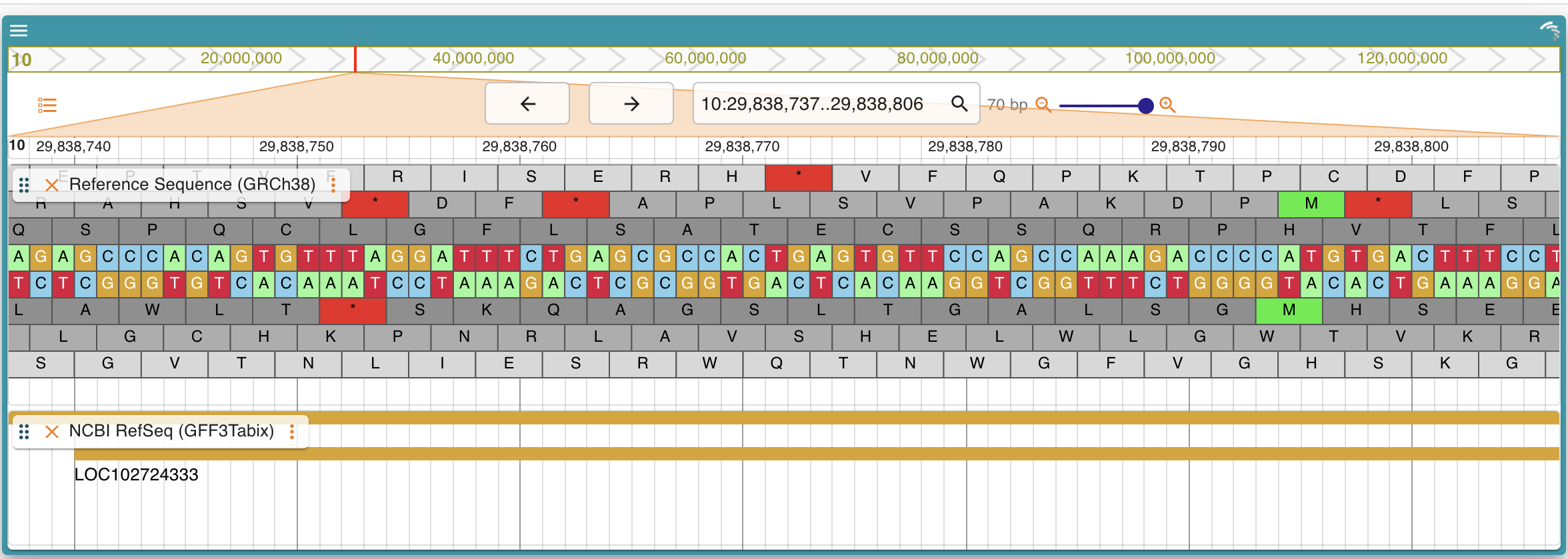
## Academic Use
This package was written with funding from the [NHGRI](https://genome.gov/) as
part of the JBrowse project. If you use it in an academic project that you
publish, please cite the most recent JBrowse paper, which will be linked from
[jbrowse.org](https://jbrowse.org/).
## Resources
- More infromation about `Dash` can be found in this [post](https://medium.com/plotly/dash-is-react-for-python-r-and-julia-c75822d1cc24)
- Examples of the `LinearGenomeView` component can be found in the [usage.py](usage.py), and [browser.py](./examples/browser.py)
- Examples of the `CircularGenomeView` component can be found in the [usage.py](usage_cgv.py)
- [JBrowse React Linear Genome View](https://jbrowse.org/storybook/lgv/main/) - documentation and examples of the React LGV component.
- [JBrowse React Circular Genome View](https://jbrowse.org/storybook/cgv/main/?path=/story/getting-started--page) - documentation and examples of the React CGV component.
- [Config Guide](https://jbrowse.org/jb2/docs/config_guide/) - a guide to configuring assemblies, tracks, text searching and more.
- [JBrowse CLI tools](https://jbrowse.org/jb2/docs/cli/) - installation and documentation
- [TextSearching](https://jbrowse.org/jb2/docs/config_guide/#text-searching) - documentation of text indexing and text search adapters
## Contact us
We **really** love talking to our users. Please reach out with any thoughts you have on what we are building!
- Report a bug or request a feature at
https://github.com/GMOD/dash_jbrowse/issues/new
- Join our developers chat at https://gitter.im/GMOD/jbrowse2
- Send an email to our mailing list at `gmod-ajax@lists.sourceforge.net`




