# [covid-seird](https://pypi.org/project/covid-seird/)
<!-- ALL-CONTRIBUTORS-BADGE:START - Do not remove or modify this section -->
[](https://pepy.tech/project/covid-seird)
[](https://opensource.org/licenses/)
[](http://hits.dwyl.com/paulorobertobranco/covid-seird)
[](https://www.codefactor.io/repository/github/paulorobertobranco/covid_seird/overview/master)

<!-- ALL-CONTRIBUTORS-BADGE:END -->
## About
**covid-seird** is a small Python package inspired by Henri Froese's post ['Infectious Disease Modelling: Beyond the Basic SIR Model'](https://towardsdatascience.com/infectious-disease-modelling-beyond-the-basic-sir-model-216369c584c4).
It implements the **SEIRD Epidemiological Model** on COVID-19 data.
- First, it **fits** a the SEIRD Model into the real timeline data of confirmed cases of COVID-19 of a country.
- Then, it **simulates** the SEIRD curves based on the previously fitted Model.
- As a result, the country's **basic reproduction number (R<sub>o</sub>)** of COVID-19 is computed.
- Also, the **fit and simulation plots** are available.
NOTE:
The COVID-19 timelines data are obtained using the [COVID19Py package](https://github.com/Kamaropoulos/COVID19Py) in order to retrieve the Worldwide Data repository operated by the Johns Hopkins University Center for Systems Science and Engineering (JHU CSSE).
## Installation
In order to install this package, simply run:
```bash
pip install covid-seird
```
## Usage
To use covid-seird, you first need to import the package and then create a new instance, passing the country code as a parameter:
```python
from covid_seird import CountryCovidSeird
brazil = CountryCovidSeird("br")
```
### Getting the country codes
```python
CountryCovidSeird.code_search("brazil")
```
output:
```python
{'BR': 'Brazil'}
```
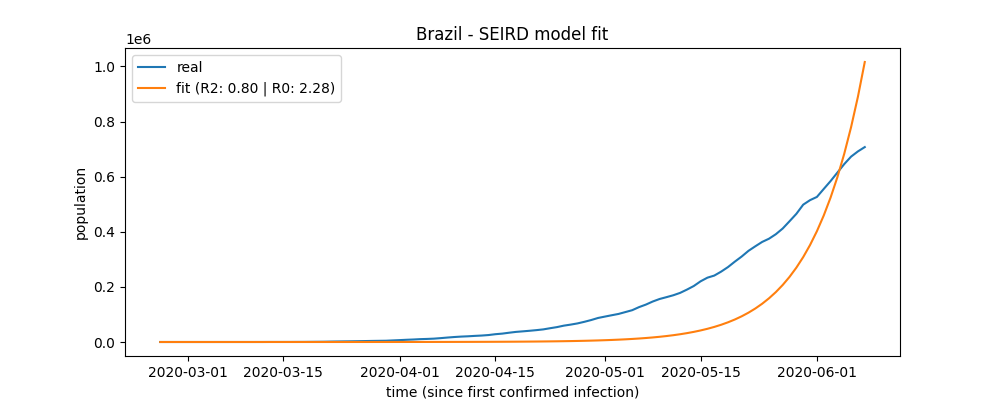
### Fit the SEIRD Model
```python
brazil.fit()
```
After the fit method's call, the fit score (R<sup>2</sup>) can be accessed:
```python
brazil.r2
```
output:
```python
0.797994368607109
```
Also after fit method's call, the basic reproduction number (R<sub>o</sub>) will be available:
```python
brazil.r0
```
output:
```python
2.280428864655168
```
### Fit plot
```python
brazil.plot_fit("brazil_fit_plot")
```
a plot file named 'brazil_fit_plot' will be created.
### SEIRD simulation
The simulation method receives as a parameter how many days ahead of the real data the SEIRD model will be computed.
```python
brazil.simulation(days_ahead=150)
```
The SEIRD curves can be accessed after the simulation method's call
```python
brazil.curves['infected']
```
output:
```python
array([4.77396851e-09, 5.32992037e-09, 5.95375686e-09, 6.59631344e-09,
7.32558927e-09, 8.14396099e-09, 9.05142859e-09, 1.00479921e-08,
...
7.48473200e-03, 7.06776956e-03, 6.67386382e-03, 6.30175876e-03,
5.95026480e-03, 5.61825534e-03, 5.30466353e-03, 5.00847963e-03])
```
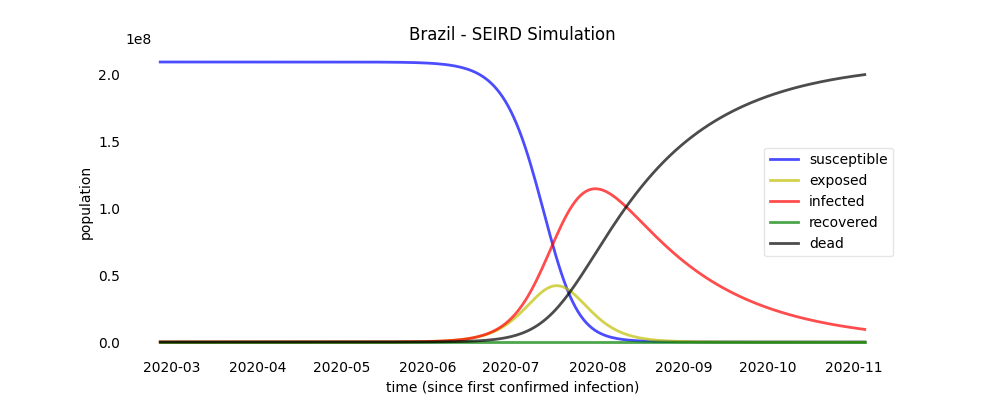
### Simulation plots
```python
brazil.plot_simulation("brazil_simulation_plot")
```
a plot file named 'brazil_simulation_plot' will be created.