<p align="center">
<img width="600" src="https://raw.githubusercontent.com/mcsorkun/ChemPlot/main/docs/logo_chemplot.png">
</p>
<br />
[](https://badge.fury.io/py/chemplot) [](https://pepy.tech/project/chemplot)
[](https://anaconda.org/conda-forge/chemplot) [](https://anaconda.org/conda-forge/chemplot)
[](https://github.com/mcsorkun/ChemPlot/blob/main/LICENSE) [](https://chemplot.readthedocs.io/en/latest/?badge=latest)
[](https://github.com/mcsorkun/ChemPlot/actions/workflows/tests.yml)
[](https://coveralls.io/github/mcsorkun/ChemPlot)
# ChemPlot
Chemplot is a python library for chemical space visualization that allows users to plot the chemical space of their molecular datasets. Chemplot contains both structural and tailored similarity algorithms to plot similar molecules together based on the needs of users. Moreover, it is easy to use even for non-experts.
## Resources
### User Manual
You can find the detailed features and examples in the following link: [User Manual](https://chemplot.readthedocs.io/en/latest/).
### Web Application
ChemPlot is also available as a web application. You can use it at the following link: [Web Application](https://share.streamlit.io/mcsorkun/chemplot-web/main/web_app_chemplot.py).
### Paper
You can find the details for the background on ChemPlot in our paper. You can download our paper at: [Paper](https://chemistry-europe.onlinelibrary.wiley.com/doi/full/10.1002/cmtd.202200005).
## Installation
There are two different options to install ChemPlot:
### Option 1: Use conda
To install ChemPlot using conda, run the following from the command line:
conda install -c conda-forge chemplot
### Option 2: Use pip
ChemPlot requires RDKit, which cannot be installed using pip. The
official RDKit installation documentation can be found
[here](http://www.rdkit.org/docs/Install.html).
After having installed RDKit, ChemPlot can be installed using pip by
running:
pip install chemplot
## How to use ChemPlot
ChemPlot is a cheminformatics tool whose purpose is to visualize subsets
of the chemical space in two dimensions. It uses the [RDKit chemistry
framework](http://www.rdkit.org), the
[scikit-learn](http://scikit-learn.org/stable/index.html) API and the
[umap-learn](https://github.com/lmcinnes/umap) API.
### Getting started
To demonstrate how to use the functions the library offers we use
[BBBP](https://github.com/mcsorkun/ChemPlot/blob/main/tests/test_data/C_2039_BBBP_2.csv) (blood-brain barrier penetration) [1] molecular dataset. BBBP is a
set of molecules encoded as SMILES, which have been assigned a binary
label according to their permeability properties. This dataset can be retrieved
from the library as a [pandas](https://pandas.pydata.org/pandas-docs/stable/index.html)
DataFrame object.
``` {.sourceCode .python3}
import chemplot as cp
data_BBBP = cp.load_data("BBBP")
```
To visualize the molecules in 2D according to their similarity it is
first needed to construct a `Plotter` object. This is the class
containing all the functions ChemPlot uses to produce the desired
visualizations. A `Plotter` object can be constructed using
classmethods, which differentiate between the type of input that is feed
to the object. In our example we need to use the method from\_smiles. We
pass three parameters: the list of SMILES from the BBBP dataset, their
target values (the binary labels) and the target type (in this case “C”,
which stands for “Classification”).
``` {.sourceCode .python3}
plotter = cp.Plotter.from_smiles(data_BBBP["smiles"], target=data_BBBP["target"], target_type="C")
```
### Plotting the results
When the `Plotter` object was constructed descriptors for each SMILES
were calculated, using the library
[mordred](http://mordred-descriptor.github.io/documentation/v0.1.0/introduction.html),
and then selected based on the target values. We reduce the number of
dimensions for each molecule from the number of descriptors selected to only 2.
ChemPlot uses three different algorithms in order to achieve this.
In this example we will first use t-SNE [2].
``` {.sourceCode .python3}
plotter.tsne()
```
The output will be a dataframe containg the reduced dimensions and the target values.
| t-SNE-1 | t-SNE-2 | target |
|------------------|------------------|------------------|
| -41.056122 | 0.355575 | 1 |
| -35.535915 | 21.648867 | 1 |
| 23.771597 | -14.438373 | 1 |
To now visualize the chemical space of the dataset we use `visualize_plot()`.
``` {.sourceCode .python3}
plotter.visualize_plot()
```
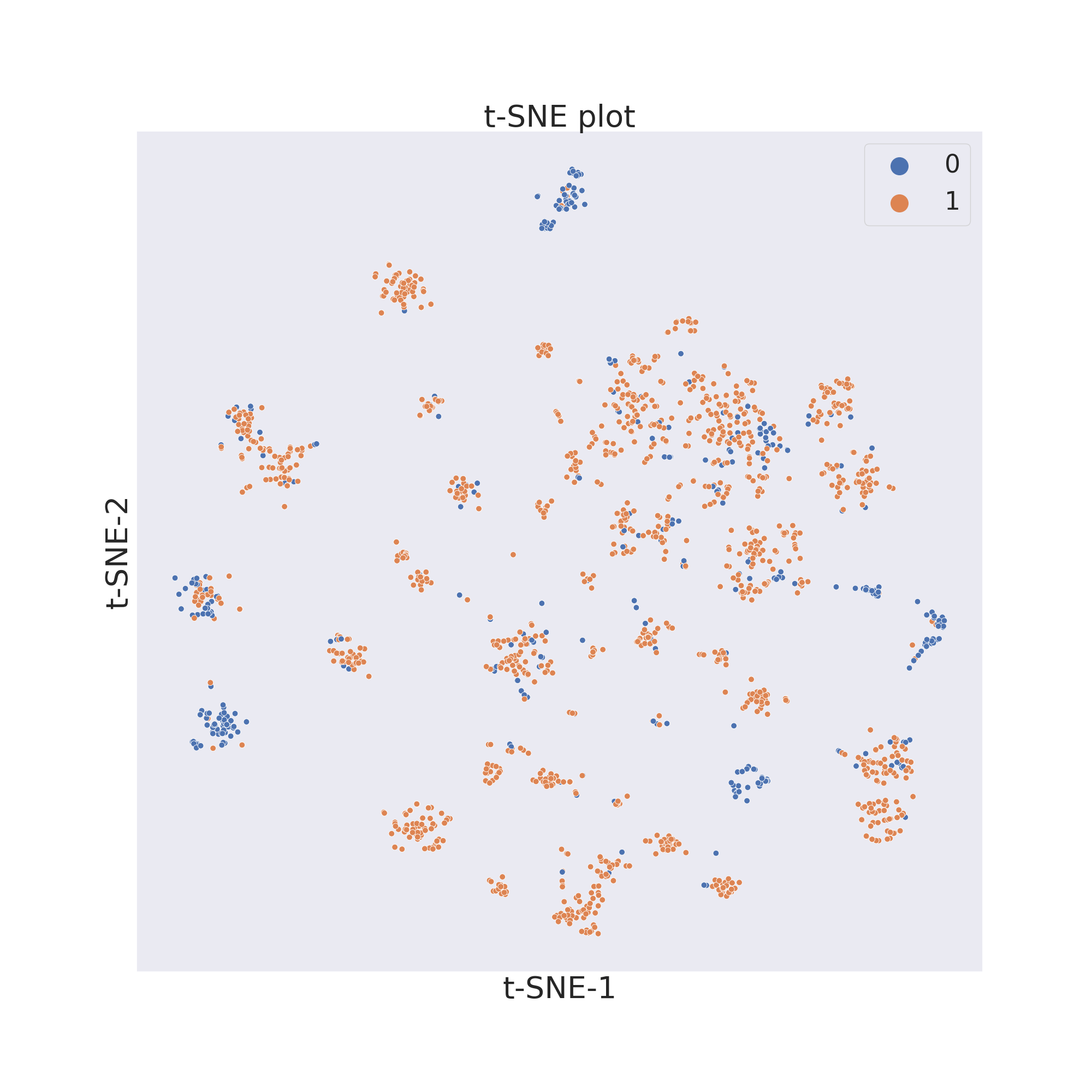
The second figure shows the results obtained by reducing the dimensions
of features Principal Component Analysis (PCA) [3].
``` {.sourceCode .python3}
plotter.pca()
plotter.visualize_plot()
```
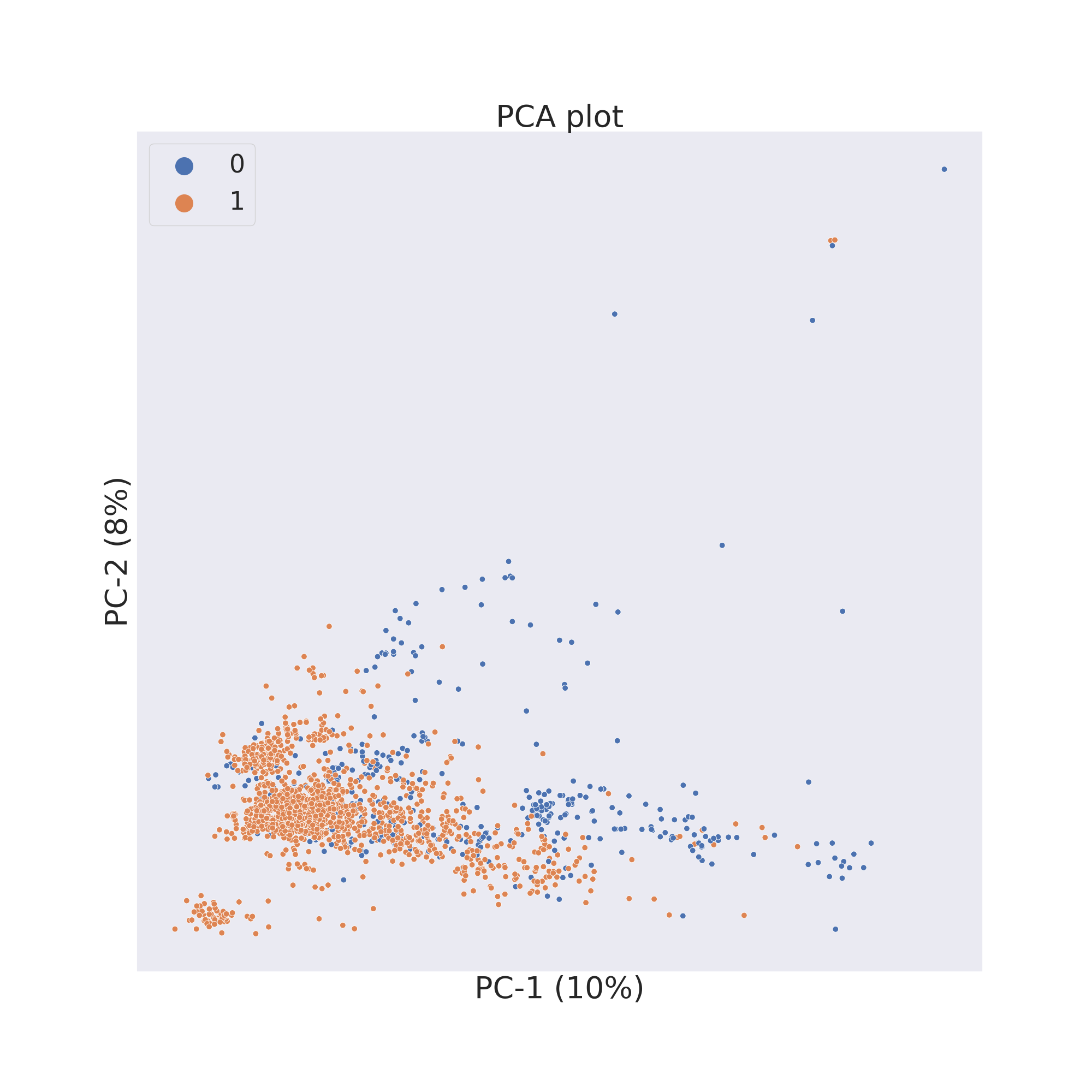
The third figure shows the results obtained by reducing the dimensions
of features by UMAP [4].
``` {.sourceCode .python3}
plotter.umap()
plotter.visualize_plot()
```
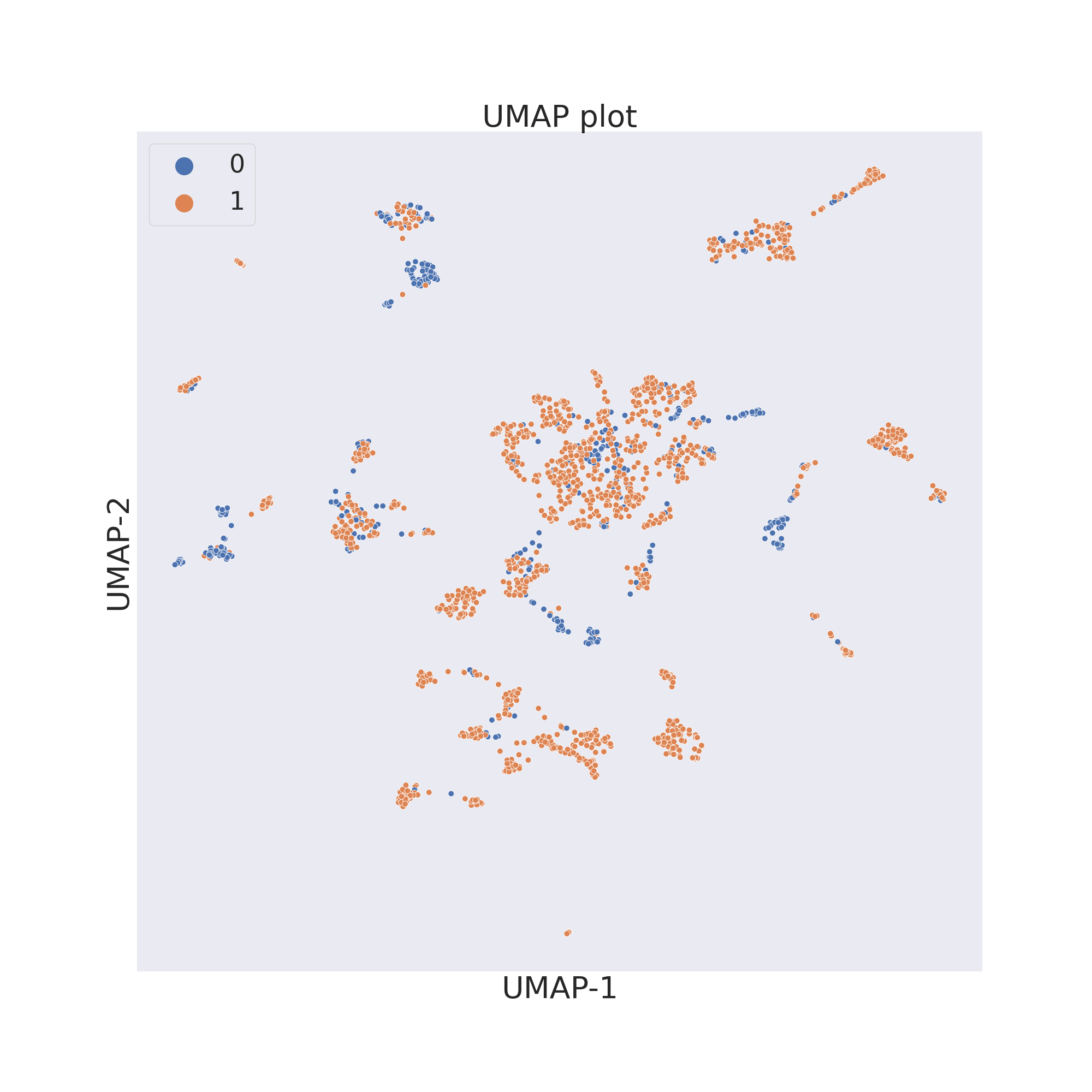
In each figure the molecules are coloured by class value.
* * * * *
<h3>
References:
</h3>
[1]: **Martins, Ines Filipa, et al.** (2012). [A Bayesian approach to
in silico blood-brain barrier penetration
modeling.](https://pubmed.ncbi.nlm.nih.gov/22612593/) Journal of
chemical information and modeling 52.6, 1686-1697
[2]: **van der Maaten, Laurens, Hinton, Geoffrey.** (2008).
[Viualizingdata using
t-SNE.](https://www.jmlr.org/papers/volume9/vandermaaten08a/vandermaaten08a.pdf?fbclid=IwAR0Bgg1eA5TFmqOZeCQXsIoL6PKrVXUFaskUKtg6yBhVXAFFvZA6yQiYx-M)
Journal of Machine Learning Research. 9. 2579-2605.
[3]: **Wold, S., Esbensen, K., Geladi, P.** (1987). [Principal
component
analysis.](https://www.sciencedirect.com/science/article/abs/pii/0169743987800849)
Chemometrics and intelligent laboratory systems. 2(1-3). 37-52.
[4]: **McInnes, L., Healy, J., Melville, J.** (2018). [Umap: Uniform
manifold approximation and projection for dimension
reduction.](https://arxiv.org/abs/1802.03426) arXivpreprint
arXiv:1802.03426.
### Contact
For any question you can contact us through email:
- [Murat Cihan Sorkun](mailto:mcsorkun@gmail.com)
- [Dajt Mullaj](mailto:dajt.mullai@gmail.com)




